Merge branch 'version-1' into feature/bd-29
Conflicts: .gitignore README.md auth/dktk dktk-fed/docker-compose.yml landing/index.html lib/generate.sh
This commit is contained in:
13
.gitignore
vendored
13
.gitignore
vendored
@@ -1,16 +1,7 @@
|
||||
##Ignore site configuration
|
||||
.gitmodules
|
||||
site-config
|
||||
|
||||
## Ignore site configuration
|
||||
config/**/*
|
||||
!config/**/*.default
|
||||
docker-compose.override.yml
|
||||
site.conf
|
||||
.bash_logout
|
||||
.bash_profile
|
||||
.bashrc
|
||||
.bash_history
|
||||
.rnd
|
||||
.pki/*
|
||||
.viminfo
|
||||
lading/*
|
||||
certs/*
|
||||
|
||||
212
README.md
212
README.md
@@ -1,14 +1,86 @@
|
||||
# Bridgehead
|
||||
|
||||
This repository contains all information and tools to deploy a bridgehead. If you have any questions about deploying a bridgehead, please [contact us](mailto:verbis-support@dkfz-heidelberg.de).
|
||||
|
||||
|
||||
TOC
|
||||
|
||||
1. [About](#about)
|
||||
- [Projects](#projects)
|
||||
- [GBA/BBMRI-ERIC](#gbabbmri-eric)
|
||||
- [DKTK/C4](#dktkc4)
|
||||
- [NNGM](#nngm)
|
||||
- [Bridgehead Components](#bridgehead-components)
|
||||
- [Blaze Server](#blaze-serverhttpsgithubcomsamplyblaze)
|
||||
1. [Requirements](#requirements)
|
||||
- [Hardware](#hardware)
|
||||
- [System](#system-requirements)
|
||||
2. [Getting Started](#getting-started)
|
||||
- [DKTK](#dktkc4)
|
||||
- [C4](#c4)
|
||||
- [GBA/BBMRI-ERIC](#gbabbmri-eric)
|
||||
3. [Configuration](#configuration)
|
||||
4. [Managing your Bridgehead](#managing-your-bridgehead)
|
||||
- [Systemd](#on-a-server)
|
||||
- [Without Systemd](#on-developers-machine)
|
||||
4. [Pitfalls](#pitfalls)
|
||||
5. [Migration-guide](#migration-guide)
|
||||
7. [License](#license)
|
||||
|
||||
|
||||
## About
|
||||
|
||||
TODO: Insert comprehensive feature list of the bridgehead? Why would anyone install it?
|
||||
|
||||
TODO: TOC
|
||||
### Projects
|
||||
|
||||
#### GBA/BBMRI-ERIC
|
||||
|
||||
The **Sample Locator** is a tool that allows researchers to make searches for samples over a large number of geographically distributed biobanks. Each biobank runs a so-called **Bridgehead** at its site, which makes it visible to the Sample Locator. The Bridgehead is designed to give a high degree of protection to patient data. Additionally, a tool called the [Negotiator][negotiator] puts you in complete control over which samples and which data are delivered to which researcher.
|
||||
|
||||
You will most likely want to make your biobanks visible via the [publicly accessible Sample Locator][sl], but the possibility also exists to install your own Sample Locator for your site or organization, see the GitHub pages for [the server][sl-server-src] and [the GUI][sl-ui-src].
|
||||
|
||||
The Bridgehead has two primary components:
|
||||
* The **Blaze Store**. This is a highly responsive FHIR data store, which you will need to fill with your data via an ETL chain.
|
||||
* The **Connector**. This is the communication portal to the Sample Locator, with specially designed features that make it possible to run it behind a corporate firewall without making any compromises on security.
|
||||
|
||||
#### DKTK/C4
|
||||
|
||||
TODO:
|
||||
|
||||
#### NNGM
|
||||
|
||||
TODO:
|
||||
|
||||
### Bridgehead Components
|
||||
|
||||
#### [Blaze Server](https://github.com/samply/blaze)
|
||||
|
||||
This holds the actual data being searched. This store must be filled by you, generally by running an ETL on your locally stored data to turn it into the standardized FHIR format that we require.
|
||||
|
||||
#### [Connector]
|
||||
|
||||
TODO:
|
||||
|
||||
|
||||
|
||||
## Requirements
|
||||
|
||||
### Hardware
|
||||
|
||||
For running your bridgehead we recommend the follwing Hardware:
|
||||
|
||||
- 4 CPU cores
|
||||
- At least 8 GB Ram
|
||||
- 10GB Hard Drive + how many data GB you need to store in the bridgehead
|
||||
|
||||
|
||||
### System Requirements
|
||||
|
||||
Before starting the installation process, please ensure that following software is available on your system:
|
||||
|
||||
### [Git](https://git-scm.com/book/en/v2/Getting-Started-Installing-Git)
|
||||
#### [Git](https://git-scm.com/book/en/v2/Getting-Started-Installing-Git)
|
||||
|
||||
To check that you have a working git installation, please run
|
||||
``` shell
|
||||
cd ~/;
|
||||
@@ -18,7 +90,8 @@ rm -rf Hello-World;
|
||||
```
|
||||
If you see the output "Hello World!" your installation should be working.
|
||||
|
||||
### [Docker](https://docs.docker.com/get-docker/)
|
||||
#### [Docker](https://docs.docker.com/get-docker/)
|
||||
|
||||
To check your docker installation, you can try to execute dockers "Hello World" Image. The command is:
|
||||
``` shell
|
||||
docker run --rm --name hello-world hello-world;
|
||||
@@ -33,7 +106,8 @@ You should also check, that the version of docker installed by you is newer than
|
||||
docker --version
|
||||
```
|
||||
|
||||
### [Docker Compose](https://docs.docker.com/compose/cli-command/#installing-compose-v2)
|
||||
#### [Docker Compose](https://docs.docker.com/compose/cli-command/#installing-compose-v2)
|
||||
|
||||
To check your docker-compose installation, please run the following command. It uses the "hello-world" image from the previous section:
|
||||
``` shell
|
||||
docker-compose -f - up <<EOF
|
||||
@@ -51,7 +125,8 @@ You should also ensure, that the version of docker-compose installed by you is n
|
||||
docker-compose --version
|
||||
```
|
||||
|
||||
### [systemd](https://systemd.io/)
|
||||
#### [systemd](https://systemd.io/)
|
||||
|
||||
You shouldn't need to install it yourself. If systemd is not available on your system you should get another system.
|
||||
To check if systemd is available on your system, please execute
|
||||
|
||||
@@ -59,6 +134,8 @@ To check if systemd is available on your system, please execute
|
||||
systemctl --version
|
||||
```
|
||||
|
||||
---
|
||||
|
||||
## Getting Started
|
||||
|
||||
If your system passed all checks from ["Requirements" section], you are now ready to download the bridgehead.
|
||||
@@ -66,14 +143,30 @@ If your system passed all checks from ["Requirements" section], you are now read
|
||||
First, clone the repository to the directory "/srv/docker/bridgehead":
|
||||
|
||||
``` shell
|
||||
sudo mkdir /srv/docker/;
|
||||
sudo mkdir -p /srv/docker/;
|
||||
sudo git clone https://github.com/samply/bridgehead.git /srv/docker/bridgehead;
|
||||
```
|
||||
|
||||
<<<<<<< HEAD
|
||||
adduser --no-create-home --disabled-login --ingroup docker --gecos "" bridgehead
|
||||
useradd -M -g docker -N -s /sbin/nologin bridgehead
|
||||
chown bridghead /srv/docker/bridgehead/ -R
|
||||
|
||||
=======
|
||||
The next step is to create a user for the bridgehead service
|
||||
|
||||
``` shell
|
||||
#!/bin/bash
|
||||
|
||||
mkdir /srv/docker && cd /srv/docker
|
||||
|
||||
adduser --no-create-home --disabled-login --ingroup docker --gecos "" bridgehead
|
||||
useradd -M -g docker -N -s /sbin/nologin bridgehead
|
||||
|
||||
chown 777 /srv/docker/bridgehead bridgehead
|
||||
sudo chown bridgehead /srv/docker/bridgehead/
|
||||
```
|
||||
>>>>>>> version-1
|
||||
|
||||
Next, you need to configure a set of variables, specific for your site with not so high security concerns. You can visit the configuration template at [GitHub](https://github.com/samply/bridgehead-config). You can download the repositories contents and add them to the "bridgehead-config" directory.
|
||||
|
||||
@@ -114,6 +207,7 @@ Environment=https_proxy=
|
||||
|
||||
|
||||
### DKTK/C4
|
||||
|
||||
You can create the site specific configuration with:
|
||||
|
||||
``` shell
|
||||
@@ -149,6 +243,7 @@ sudo systemctl bridgehead@dktk.service;
|
||||
```
|
||||
|
||||
### C4
|
||||
|
||||
You can create the site specific configuration with:
|
||||
|
||||
``` shell
|
||||
@@ -212,12 +307,30 @@ sudo systemctl bridgehead@gbn.service;
|
||||
```
|
||||
|
||||
### Developers
|
||||
|
||||
Because some developers machines doesn't support system units (e.g Windows Subsystem for Linux), we provide a dev environment [configuration script](./lib/init-test-environment.sh).
|
||||
It is not recommended to use this script in production!
|
||||
|
||||
## Configuration
|
||||
|
||||
### Basic Auth
|
||||
|
||||
use add_user.sh
|
||||
|
||||
|
||||
### HTTPS Access
|
||||
|
||||
We advise to use https for all service of your bridgehead. HTTPS is enabled on default. For starting the bridghead you need a ssl certificate. You can either create it yourself or get a signed one. You need to drop the certificates in /certs.
|
||||
|
||||
If you want to create it yourself, you can generate the necessary certs with:
|
||||
|
||||
``` shell
|
||||
openssl req -x509 -newkey rsa:4096 -nodes -keyout certs/traefik.key -out certs/traefik.crt -days 365
|
||||
```
|
||||
|
||||
|
||||
### Locally Managed Secrets
|
||||
|
||||
This section describes the secrets you need to configure locally through the configuration
|
||||
|
||||
| Name | Recommended Value | Description |
|
||||
@@ -240,55 +353,77 @@ This section describes the secrets you need to configure locally through the con
|
||||
| MAGICPL_OIDC_CLIENT_SECRET || The client secret used for your machine, to connect with the central authentication service |
|
||||
|
||||
### Cooperatively Managed Secrets
|
||||
|
||||
> TODO: Describe secrets from site-config
|
||||
|
||||
## Managing your Bridgehead
|
||||
|
||||
> TODO: Rewrite this section (restart, stop, uninstall, manual updates)
|
||||
|
||||
### On a Server
|
||||
|
||||
#### Start
|
||||
|
||||
This will start a not running bridgehead system unit:
|
||||
``` shell
|
||||
sudo systemctl start bridgehead@<dktk/c4/gbn>
|
||||
```
|
||||
|
||||
#### Stop
|
||||
|
||||
This will stop a running bridgehead system unit:
|
||||
``` shell
|
||||
sudo systemctl stop bridgehead@<dktk/c4/gbn>
|
||||
```
|
||||
|
||||
#### Update
|
||||
|
||||
This will update bridgehead system unit:
|
||||
``` shell
|
||||
sudo systemctl start bridgehead-update@<dktk/c4/gbn>
|
||||
```
|
||||
|
||||
#### Remove the Bridgehead System Units
|
||||
|
||||
If, for some reason you want to remove the installed bridgehead units, we added a [script](./lib/remove-bridgehead-units.sh) you can execute:
|
||||
``` shell
|
||||
sudo ./lib/remove-bridgehead-units.sh
|
||||
```
|
||||
|
||||
### On Developers Machine
|
||||
|
||||
For developers, we provide additional scripts for starting and stopping the specif bridgehead:
|
||||
|
||||
#### Start
|
||||
|
||||
This shell script start a specified bridgehead. Choose between "dktk", "c4" and "gbn".
|
||||
``` shell
|
||||
./start-bridgehead <dktk/c4/gbn>
|
||||
```
|
||||
|
||||
#### Stop
|
||||
|
||||
This shell script stops a specified bridgehead. Choose between "dktk", "c4" and "gbn".
|
||||
``` shell
|
||||
./stop-bridgehead <dktk/c4/gbn>
|
||||
```
|
||||
|
||||
#### Update
|
||||
|
||||
This shell script updates the configuration for all bridgeheads installed on your system.
|
||||
``` shell
|
||||
./update-bridgehead
|
||||
```
|
||||
> NOTE: If you want to regularly update your developing instance, you can create a CRON job that executes this script.
|
||||
|
||||
## Migration Guide
|
||||
|
||||
> TODO: How to transfer from windows/gbn
|
||||
|
||||
## Pitfalls
|
||||
|
||||
### [Git Proxy Configuration](https://gist.github.com/evantoli/f8c23a37eb3558ab8765)
|
||||
|
||||
Unlike most other tools, git doesn't use the default proxy variables "http_proxy" and "https_proxy". To make git use a proxy, you will need to adjust the global git configuration:
|
||||
|
||||
``` shell
|
||||
@@ -305,6 +440,7 @@ sudo git config --global --list;
|
||||
```
|
||||
|
||||
### Docker Daemon Proxy Configuration
|
||||
|
||||
Docker has a background daemon, responsible for downloading images and starting them. To configure the proxy for this daemon, use the systemctl command:
|
||||
|
||||
``` shell
|
||||
@@ -332,3 +468,67 @@ To make the configuration effective, you need to tell systemd to reload the conf
|
||||
sudo systemctl daemon-reload;
|
||||
sudo systemctl restart docker;
|
||||
```
|
||||
|
||||
## After the Installtion
|
||||
|
||||
After starting your bridgehead, visit the landing page under the hostname. If you singed your own ssl certificate, there is probable an error message. However, you can accept it as exception.
|
||||
|
||||
On this page, there are all important links to each component, central and local.
|
||||
|
||||
### Connector Administration
|
||||
|
||||
The Connector administration panel allows you to set many of the parameters regulating your Bridgehead. Most especially, it is the place where you can register your site with the Sample Locator. To access this page, proceed as follows:
|
||||
|
||||
* Open the Connector page: https://<hostname>/<project>-connector/
|
||||
* In the "Local components" box, click the "Samply Share" button.
|
||||
* A new page will be opened, where you will need to log in using the administrator credentials (admin/adminpass by default).
|
||||
* After log in, you will be taken to the administration dashboard, allowing you to configure the Connector.
|
||||
* If this is the first time you have logged in as an administrator, you are strongly recommended to set a more secure password! You can use the "Users" button on the dashboard to do this.
|
||||
|
||||
### GBA/BBMRI-ERIC
|
||||
|
||||
#### Register with a Directory
|
||||
|
||||
The [Directory][directory] is a BBMRI project that aims to catalog all biobanks in Europe and beyond. Each biobank is given its own unique ID and the Directory maintains counts of the number of donors and the number of samples held at each biobank. You are strongly encouraged to register with the Directory, because this opens the door to further services, such as the [Negotiator][negotiator].
|
||||
|
||||
Generally, you should register with the BBMRI national node for the country where your biobank is based. You can find a list of contacts for the national nodes [here](http://www.bbmri-eric.eu/national-nodes/). If your country is not in this list, or you have any questions, please contact the [BBMRI helpdesk](mailto:directory@helpdesk.bbmri-eric.eu). If your biobank is for COVID samples, you can also take advantage of an accelerated registration process [here](https://docs.google.com/forms/d/e/1FAIpQLSdIFfxADikGUf1GA0M16J0HQfc2NHJ55M_E47TXahju5BlFIQ).
|
||||
|
||||
Your national node will give you detailed instructions for registering, but for your information, here are the basic steps:
|
||||
|
||||
* Log in to the Directory for your country.
|
||||
* Add your biobank and enter its details, including contact information for a person involved in running the biobank.
|
||||
* You will need to create at least one collection.
|
||||
* Note the biobank ID and the collection ID that you have created - these will be needed when you register with the Locator (see below).
|
||||
|
||||
#### Register with a Locator
|
||||
|
||||
* Go to the registration page http://localhost:8082/admin/broker_list.xhtml.
|
||||
* To register with a Locator, enter the following values in the three fields under "Join new Searchbroker":
|
||||
* "Address": Depends on which Locator you want to register with:
|
||||
* `https://locator.bbmri-eric.eu/broker/`: BBMRI Locator production service (European).
|
||||
* `http://147.251.124.125:8088/broker/`: BBMRI Locator test service (European).
|
||||
* `https://samplelocator.bbmri.de/broker/`: GBA Sample Locator production service (German).
|
||||
* `https://samplelocator.test.bbmri.de/broker/`: GBA Sample Locator test service (German).
|
||||
* "Your email address": this is the email to which the registration token will be returned.
|
||||
* "Automatic reply": Set this to be `Total Size`
|
||||
* Click "Join" to start the registration process.
|
||||
* You should now have a list containing exactly one broker. You will notice that the "Status" box is empty.
|
||||
* Send an email to `feedback@germanbiobanknode.de` and let us know which of our Sample Locators you would like to register to. Please include the biobank ID and the collection ID from your Directory registration, if you have these available.
|
||||
* We will send you a registration token per email.
|
||||
* You will then re-open the Connector and enter the token into the "Status" box.
|
||||
* You should send us an email to let us know that you have done this.
|
||||
* We will then complete the registration process
|
||||
* We will email you to let you know that your biobank is now visible in the Sample Locator.
|
||||
|
||||
If you are a Sample Locator administrator, you will need to understand the [registration process](./SampleLocatorRegistration.md). Normal bridgehead admins do not need to worry about this.
|
||||
|
||||
|
||||
## License
|
||||
|
||||
Copyright 2019 - 2022 The Samply Community
|
||||
|
||||
Licensed under the Apache License, Version 2.0 (the "License"); you may not use this file except in compliance with the License. You may obtain a copy of the License at
|
||||
|
||||
http://www.apache.org/licenses/LICENSE-2.0
|
||||
|
||||
Unless required by applicable law or agreed to in writing, software distributed under the License is distributed on an "AS IS" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. See the License for the specific language governing permissions and limitations under the License.
|
||||
|
||||
@@ -1,2 +0,0 @@
|
||||
patrick:$2y$05$9tYlNuZEfCi1FrSUMYM0iOz8FEsHjg3QiPpr3ZfChL81rZ8IrZ0gK
|
||||
|
||||
@@ -1,159 +0,0 @@
|
||||
version: "3.7"
|
||||
volumes:
|
||||
c4-connector-db-data:
|
||||
c4-connector-logs:
|
||||
patientlist-db-data:
|
||||
patientlist-logs:
|
||||
id-manager-logs:
|
||||
c4-store-db-data:
|
||||
c4-store-logs:
|
||||
|
||||
services:
|
||||
traefik:
|
||||
image: traefik:2.4
|
||||
command:
|
||||
- --api.insecure=true
|
||||
- --entrypoints.web.address=:80
|
||||
- --entrypoints.web-secure.address=:443
|
||||
- --providers.docker=true
|
||||
ports:
|
||||
- 80:80
|
||||
- 443:443
|
||||
- 8080:8080
|
||||
volumes:
|
||||
- /var/run/docker.sock:/var/run/docker.sock:ro
|
||||
|
||||
landing:
|
||||
image: nginx:stable
|
||||
volumes:
|
||||
- ../landing/:/usr/share/nginx/html
|
||||
labels:
|
||||
- "traefik.enable=true"
|
||||
- "traefik.http.routers.landing.rule=PathPrefix(`/`)"
|
||||
- "traefik.http.services.landing.loadbalancer.server.port=80"
|
||||
|
||||
c4-connector:
|
||||
image: "samply/share-client:c4-feature-environmentPreconfigurationTorben"
|
||||
environment:
|
||||
POSTGRES_HOST: "c4-connector-db"
|
||||
ID_MANAGER_APIKEY: ${MAGICPL_API_KEY_CONNECTOR}
|
||||
POSTGRES_PASSWORD: ${CONNECTOR_POSTGRES_PASS}
|
||||
HTTP_PROXY_USER: ${HTTP_PROXY_USER}
|
||||
HTTP_PROXY_PASSWORD: ${HTTP_PROXY_PASSWORD}
|
||||
HTTPS_PROXY_USER: ${HTTPS_PROXY_USER}
|
||||
HTTPS_PROXY_PASSWORD: ${HTTPS_PROXY_PASSWORD}
|
||||
LDM_URL: "${PROTOCOL}://${HOST}/c4-localdatamanagement"
|
||||
POSTGRES_PASSWORD: ${CONNECTOR_POSTGRES_PASS}
|
||||
env_file:
|
||||
- ../site-config/c4.env
|
||||
# Necessary for the connector to successful check the status of other components on the same host
|
||||
extra_hosts:
|
||||
- "host.docker.internal:host-gateway"
|
||||
- "${HOST}:${HOSTIP}"
|
||||
volumes:
|
||||
- "c4-connector-logs:/usr/local/tomcat/logs"
|
||||
depends_on:
|
||||
- connector-db
|
||||
restart: always
|
||||
labels:
|
||||
- "traefik.enable=true"
|
||||
- "traefik.http.routers.c4_connector.rule=PathPrefix(`/c4-connector`)"
|
||||
- "traefik.http.services.c4_connector.loadbalancer.server.port=8080"
|
||||
|
||||
c4-connector-db:
|
||||
image: postgres:10.17
|
||||
environment:
|
||||
POSTGRES_DB: "share_v2"
|
||||
POSTGRES_USER: "samplyweb"
|
||||
POSTGRES_PASSWORD: ${CONNECTOR_POSTGRES_PASS}
|
||||
volumes:
|
||||
- "c4-connector-db-data:/var/lib/postgresql/data"
|
||||
restart: always
|
||||
|
||||
id-manager:
|
||||
image: docker.verbis.dkfz.de/ccp/idmanager:bridgehead-develop
|
||||
environment:
|
||||
MAGICPL_SITE: ${SITE}
|
||||
MAGICPL_MAINZELLISTE_API_KEY: ${MAGICPL_MAINZELLISTE_API_KEY}
|
||||
MAGICPL_API_KEY: ${MAGICPL_API_KEY}
|
||||
MAGICPL_API_KEY_CONNECTOR: ${MAGICPL_API_KEY_CONNECTOR}
|
||||
MAGICPL_MAINZELLISTE_CENTRAL_API_KEY: ${MAGICPL_MAINZELLISTE_CENTRAL_API_KEY}
|
||||
MAGICPL_CENTRAL_API_KEY: ${MAGICPL_CENTRAL_API_KEY}
|
||||
MAGICPL_OIDC_CLIENT_ID: ${MAGICPL_OIDC_CLIENT_ID}
|
||||
MAGICPL_OIDC_CLIENT_SECRET: ${MAGICPL_OIDC_CLIENT_SECRET}
|
||||
TOMCAT_REVERSEPROXY_FQDN: "${HOST}"
|
||||
HTTP_PROXY_USER: ${HTTP_PROXY_USER}
|
||||
HTTP_PROXY_PASSWORD: ${HTTP_PROXY_PASSWORD}
|
||||
HTTPS_PROXY_USER: ${HTTPS_PROXY_USER}
|
||||
HTTPS_PROXY_PASSWORD: ${HTTPS_PROXY_PASSWORD}
|
||||
env_file:
|
||||
- ../site-config/dktk.env
|
||||
volumes:
|
||||
- "id-manager-logs:/usr/local/tomcat/logs"
|
||||
depends_on:
|
||||
- patientlist
|
||||
labels:
|
||||
- "traefik.http.routers.idmanager.rule=PathPrefix(`/ID-Manager`)"
|
||||
- "traefik.http.services.idmanager.loadbalancer.server.port=8080"
|
||||
|
||||
patientlist:
|
||||
image: docker.verbis.dkfz.de/ccp/patientlist:bridgehead-develop
|
||||
environment:
|
||||
ML_SITE: ${SITE}
|
||||
ML_API_KEY: ${MAGICPL_MAINZELLISTE_API_KEY}
|
||||
ML_DB_PASS: ${ML_DB_PASS}
|
||||
TOMCAT_REVERSEPROXY_FQDN: "${HOST}"
|
||||
env_file:
|
||||
- ../site-config/dktk.env
|
||||
# TODO: Implement automatic seed generation in mainzelliste
|
||||
- ../site-config/patientlist.env
|
||||
volumes:
|
||||
- "patientlist-logs:/usr/local/tomcat/logs"
|
||||
labels:
|
||||
- "traefik.http.routers.patientlist.rule=PathPrefix(`/Patientlist`)"
|
||||
- "traefik.http.services.patientlist.loadbalancer.server.port=8080"
|
||||
depends_on:
|
||||
- patientlist-db
|
||||
|
||||
patientlist-db:
|
||||
image: postgres:13.1-alpine
|
||||
environment:
|
||||
POSTGRES_DB: mainzelliste
|
||||
POSTGRES_USER: mainzelliste
|
||||
POSTGRES_PASSWORD: ${ML_DB_PASS}
|
||||
TZ: "Europe/Berlin"
|
||||
volumes:
|
||||
- "patientlist-db-data:/var/lib/postgresql/data"
|
||||
|
||||
c4-store:
|
||||
image: docker.verbis.dkfz.de/ccp/samply.store:release-5.1.2
|
||||
environment:
|
||||
MDR_NAMESPACE: "adt,dktk,marker"
|
||||
MDR_VALIDATION: false
|
||||
DEPLOYMENT_CONTEXT: "c4-localdatamanagement"
|
||||
POSTGRES_HOST: c4-store-db
|
||||
POSTGRES_PORT: 5432
|
||||
POSTGRES_DB: samplystore
|
||||
POSTGRES_USER: samplystore
|
||||
POSTGRES_PASSWORD: ${STORE_POSTGRES_PASS}
|
||||
TZ: "Europe/Berlin"
|
||||
volumes:
|
||||
- "c4-store-logs:/usr/local/tomcat/logs"
|
||||
labels:
|
||||
- "traefik.enable=true"
|
||||
- "traefik.http.routers.store_c4.rule=PathPrefix(`/c4-localdatamanagement`)"
|
||||
depends_on:
|
||||
- store-db
|
||||
restart: always
|
||||
|
||||
c4-store-db:
|
||||
image: postgres:9.5-alpine
|
||||
command: postgres -c datestyle='iso, dmy'
|
||||
environment:
|
||||
TZ: "Europe/Berlin"
|
||||
POSTGRES_DB: samplystore
|
||||
POSTGRES_USER: samplystore
|
||||
POSTGRES_PASSWORD: ${STORE_POSTGRES_PASS}
|
||||
volumes:
|
||||
- "c4-store-db-data:/var/lib/postgresql/data"
|
||||
restart: always
|
||||
110
ccp/docker-compose.yml
Normal file
110
ccp/docker-compose.yml
Normal file
@@ -0,0 +1,110 @@
|
||||
version: "3.7"
|
||||
|
||||
services:
|
||||
### Does not need proxy settings
|
||||
traefik:
|
||||
container_name: bridgehead-traefik
|
||||
image: traefik:2.4
|
||||
command:
|
||||
- --entrypoints.web.address=:80
|
||||
- --entrypoints.websecure.address=:443
|
||||
- --providers.docker=true
|
||||
- --api.dashboard=true
|
||||
- --accesslog=true # print access-logs
|
||||
- --entrypoints.web.http.redirections.entrypoint.to=websecure
|
||||
- --entrypoints.web.http.redirections.entrypoint.scheme=https
|
||||
labels:
|
||||
- "traefik.http.routers.dashboard.rule=PathPrefix(`/api`) || PathPrefix(`/dashboard`)"
|
||||
- "traefik.http.routers.dashboard.entrypoints=websecure"
|
||||
- "traefik.http.routers.dashboard.service=api@internal"
|
||||
- "traefik.http.routers.dashboard.tls=true"
|
||||
- "traefik.http.routers.dashboard.middlewares=auth"
|
||||
- "traefik.http.middlewares.auth.basicauth.users=${bc_auth_users}"
|
||||
ports:
|
||||
- 80:80
|
||||
- 443:443
|
||||
volumes:
|
||||
- ../certs:/tools/certs
|
||||
- /var/run/docker.sock:/var/run/docker.sock:ro
|
||||
extra_hosts:
|
||||
- "host.docker.internal:host-gateway"
|
||||
|
||||
|
||||
### Does need to know the outside proxy to connect central components
|
||||
forward_proxy:
|
||||
container_name: bridgehead-squid
|
||||
image: ubuntu/squid
|
||||
environment:
|
||||
http_proxy: ${http_proxy}
|
||||
https_proxy: ${https_proxy}
|
||||
volumes:
|
||||
- "bridgehead-proxy:/var/log/squid"
|
||||
|
||||
## Needs internal proxy config
|
||||
landing:
|
||||
container_name: bridgehead-landingpage
|
||||
image: nginx:stable
|
||||
volumes:
|
||||
- ../landing/:/usr/share/nginx/html
|
||||
labels:
|
||||
- "traefik.enable=true"
|
||||
- "traefik.http.routers.landing.rule=PathPrefix(`/`)"
|
||||
- "traefik.http.services.landing.loadbalancer.server.port=80"
|
||||
- "traefik.http.routers.landing.tls=true"
|
||||
|
||||
## Needs internal proxy config
|
||||
blaze:
|
||||
image: "samply/blaze:0.17"
|
||||
container_name: bridgehead-ccp-blaze
|
||||
environment:
|
||||
BASE_URL: "http://blaze:8080"
|
||||
JAVA_TOOL_OPTIONS: "-Xmx4g"
|
||||
LOG_LEVEL: "debug"
|
||||
ENFORCE_REFERENTIAL_INTEGRITY: "false"
|
||||
volumes:
|
||||
- "blaze-data:/app/data"
|
||||
labels:
|
||||
- "traefik.enable=true"
|
||||
- "traefik.http.middlewares.test-auth.basicauth.users=${bc_auth_users}"
|
||||
- "traefik.http.routers.blaze_ccp.rule=PathPrefix(`/ccp-localdatamanagement`)"
|
||||
- "traefik.http.middlewares.ccp_b_strip.stripprefix.prefixes=/ccp-localdatamanagement"
|
||||
- "traefik.http.services.blaze_ccp.loadbalancer.server.port=8080"
|
||||
- "traefik.http.routers.blaze_ccp.middlewares=ccp_b_strip,test-auth"
|
||||
- "traefik.http.routers.blaze_ccp.tls=true"
|
||||
|
||||
ccp-search-share:
|
||||
image: "ghcr.io/samply/dktk-fed-search-share:main"
|
||||
container_name: bridgehead-ccp-share
|
||||
environment:
|
||||
APP_BASE_URL: "http://dktk-fed-search-share:8080"
|
||||
APP_BROKER_BASEURL: "https://dktk-fed-search.verbis.dkfz.de/broker/rest/searchbroker"
|
||||
APP_BROKER_MAIL: "foo@bar.de"
|
||||
APP_STORE_BASEURL: "http://bridgehead-ccp-blaze:8080/fhir"
|
||||
SPRING_DATASOURCE_URL: "jdbc:postgresql://bridgehead-ccp-share-db:5432/dktk-fed-search-share"
|
||||
JAVA_TOOL_OPTIONS: "-Xmx1g"
|
||||
http_proxy: "http://bridgehead-squid:3128"
|
||||
https_proxy: "http://bridgehead-squid:3128"
|
||||
HTTP_PROXY: "http://bridgehead-squid:3128"
|
||||
HTTPS_PROXY: "http://bridgehead-squid:3128"
|
||||
depends_on:
|
||||
- ccp-search-share-db
|
||||
- blaze
|
||||
labels:
|
||||
- "traefik.enable=true"
|
||||
- "traefik.http.routers.dktk-fed-search.rule=PathPrefix(`/cpp-connector`)"
|
||||
- "traefik.http.services.dktk-fed-search.loadbalancer.server.port=8080"
|
||||
|
||||
ccp-search-share-db:
|
||||
image: "postgres:14"
|
||||
container_name: bridgehead-ccp-share-db
|
||||
environment:
|
||||
POSTGRES_USER: "dktk-fed-search-share"
|
||||
POSTGRES_PASSWORD: "dktk-fed-search-share"
|
||||
POSTGRES_DB: "dktk-fed-search-share"
|
||||
volumes:
|
||||
- "ccp-search-share-db-data:/var/lib/postgresql/data"
|
||||
|
||||
volumes:
|
||||
blaze-data:
|
||||
bridgehead-proxy:
|
||||
ccp-search-share-db-data:
|
||||
@@ -1,91 +0,0 @@
|
||||
version: "3.7"
|
||||
services:
|
||||
traefik:
|
||||
container_name: bridgehead_traefik
|
||||
image: traefik:2.4
|
||||
command:
|
||||
- --api.insecure=true
|
||||
- --entrypoints.web.address=:80
|
||||
- --entrypoints.websecure.address=:443
|
||||
- --providers.docker=true
|
||||
- --providers.file.directory=/configuration/
|
||||
- --entrypoints.web.http.redirections.entrypoint.to=websecure
|
||||
- --entrypoints.web.http.redirections.entrypoint.scheme=https
|
||||
- --providers.file.watch=true
|
||||
ports:
|
||||
- 80:80
|
||||
- 443:443
|
||||
- 8080:8080
|
||||
volumes:
|
||||
- ../certs:/tools/certs
|
||||
- ../tools/traefik/:/configuration/
|
||||
- /var/run/docker.sock:/var/run/docker.sock:ro
|
||||
- ../auth/:/auth
|
||||
extra_hosts:
|
||||
- "host.docker.internal:host-gateway"
|
||||
|
||||
landing:
|
||||
container_name: bridgehead_landingpage
|
||||
image: nginx:stable
|
||||
volumes:
|
||||
- ../landing/:/usr/share/nginx/html
|
||||
labels:
|
||||
- "traefik.enable=true"
|
||||
- "traefik.http.routers.landing.rule=PathPrefix(`/`)"
|
||||
- "traefik.http.services.landing.loadbalancer.server.port=80"
|
||||
- "traefik.http.routers.landing.tls=true"
|
||||
|
||||
blaze:
|
||||
image: "samply/blaze:0.16"
|
||||
container_name: bridgehead_dktk_blaze
|
||||
environment:
|
||||
BASE_URL: "http://blaze:8080"
|
||||
JAVA_TOOL_OPTIONS: "-Xmx4g"
|
||||
LOG_LEVEL: "debug"
|
||||
ENFORCE_REFERENTIAL_INTEGRITY: "false"
|
||||
volumes:
|
||||
- "blaze-data:/app/data"
|
||||
labels:
|
||||
- "traefik.enable=true"
|
||||
- "traefik.http.middlewares.test-auth.basicauth.usersfile=/auth/dktk"
|
||||
- "traefik.http.routers.blaze_dktk.rule=PathPrefix(`/dktk-localdatamanagement`)"
|
||||
- "traefik.http.middlewares.dktk_b_strip.stripprefix.prefixes=/dktk-localdatamanagement"
|
||||
- "traefik.http.services.blaze_dktk.loadbalancer.server.port=8080"
|
||||
- "traefik.http.routers.blaze_dktk.middlewares=dktk_b_strip,test-auth"
|
||||
- "traefik.http.routers.blaze_dktk.tls=true"
|
||||
|
||||
# dktk-fed-search-share:
|
||||
# image: "ghcr.io/samply/dktk-fed-search-share:pr-1"
|
||||
# container_name: bridgehead_dktk_share
|
||||
# environment:
|
||||
# APP_BASE_URL: "http://dktk-fed-search-share:8080"
|
||||
# APP_STORE_URL: "http://blaze:8080/fhir"
|
||||
# APP_BROKER_BASEURL: "http://e260-serv-11.inet.dkfz-heidelberg.de:8080/broker/rest/searchbroker"
|
||||
# APP_BROKER_MAIL: "foo@bar.de"
|
||||
# APP_STORE_BASEURL: "http://bridgehead_dktk_blaze:8080/fhir"
|
||||
# SPRING_DATASOURCE_URL: "jdbc:postgresql://dktk-fed-search-share-db:5432/dktk-fed-search-share"
|
||||
# JAVA_TOOL_OPTIONS: "-Xmx1g"
|
||||
# http_proxy: "http://www-int2.inet.dkfz-heidelberg.de:3128"
|
||||
# https_proxy: "http://www-int2.inet.dkfz-heidelberg.de:3128"
|
||||
# HTTP_PROXY: "http://www-int2.inet.dkfz-heidelberg.de:3128"
|
||||
# HTTPS_PROXY: "http://www-int2.inet.dkfz-heidelberg.de:3128"
|
||||
# depends_on:
|
||||
# - dktk-fed-search-share-db
|
||||
# - blaze
|
||||
# labels:
|
||||
# - "traefik.enable=true"
|
||||
# - "traefik.http.routers.dktk-fed-search.rule=PathPrefix(`/dktk-connector`)"
|
||||
# - "traefik.http.services.dktk-fed-search.loadbalancer.server.port=8080"
|
||||
|
||||
# dktk-fed-search-share-db:
|
||||
# image: "postgres:14"
|
||||
# environment:
|
||||
# POSTGRES_USER: "dktk-fed-search-share"
|
||||
# POSTGRES_PASSWORD: "dktk-fed-search-share"
|
||||
# POSTGRES_DB: "dktk-fed-search-share"
|
||||
# volumes:
|
||||
# - "dktk-fed-search-share-db-data:/var/lib/postgresql/data"
|
||||
|
||||
volumes:
|
||||
blaze-data:
|
||||
# dktk-fed-search-share-db-data:
|
||||
@@ -1,122 +0,0 @@
|
||||
version: "3.7"
|
||||
volumes:
|
||||
dktk-connector-db-data:
|
||||
dktk-connector-logs:
|
||||
patientlist-db-data:
|
||||
patientlist-logs:
|
||||
id-manager-logs:
|
||||
|
||||
services:
|
||||
traefik:
|
||||
image: traefik:2.4
|
||||
command:
|
||||
- --api.insecure=true
|
||||
- --entrypoints.web.address=:80
|
||||
- --entrypoints.web-secure.address=:443
|
||||
- --providers.docker=true
|
||||
ports:
|
||||
- 80:80
|
||||
- 443:443
|
||||
- 8080:8080
|
||||
volumes:
|
||||
- /var/run/docker.sock:/var/run/docker.sock:ro
|
||||
|
||||
landing:
|
||||
image: nginx:stable
|
||||
volumes:
|
||||
- ../landing/:/usr/share/nginx/html
|
||||
labels:
|
||||
- "traefik.enable=true"
|
||||
- "traefik.http.routers.landing.rule=PathPrefix(`/`)"
|
||||
- "traefik.http.services.landing.loadbalancer.server.port=80"
|
||||
|
||||
dktk-connector:
|
||||
image: "samply/share-client:dktk-feature-environmentPreconfigurationTorben"
|
||||
environment:
|
||||
POSTGRES_HOST: "dktk-connector-db"
|
||||
ID_MANAGER_APIKEY: ${MAGICPL_API_KEY_CONNECTOR}
|
||||
POSTGRES_PASSWORD: ${CONNECTOR_POSTGRES_PASS}
|
||||
HTTP_PROXY_USER: ${HTTP_PROXY_USER}
|
||||
HTTP_PROXY_PASSWORD: ${HTTP_PROXY_PASSWORD}
|
||||
HTTPS_PROXY_USER: ${HTTPS_PROXY_USER}
|
||||
HTTPS_PROXY_PASSWORD: ${HTTPS_PROXY_PASSWORD}
|
||||
env_file:
|
||||
- ../site-config/dktk.env
|
||||
# Necessary for the connector to successful check the status of other components on the same host
|
||||
extra_hosts:
|
||||
- "host.docker.internal:host-gateway"
|
||||
- "${HOST}:${HOSTIP}"
|
||||
volumes:
|
||||
- "dktk-connector-logs:/usr/local/tomcat/logs"
|
||||
depends_on:
|
||||
- connector-db
|
||||
restart: always
|
||||
labels:
|
||||
- "traefik.enable=true"
|
||||
- "traefik.http.routers.dktk_connector.rule=PathPrefix(`/dktk-connector`)"
|
||||
- "traefik.http.services.dktk_connector.loadbalancer.server.port=8080"
|
||||
|
||||
dktk-connector-db:
|
||||
image: postgres:10.17
|
||||
environment:
|
||||
POSTGRES_DB: "share_v2"
|
||||
POSTGRES_USER: "samplyweb"
|
||||
POSTGRES_PASSWORD: ${CONNECTOR_POSTGRES_PASS}
|
||||
volumes:
|
||||
- "dktk-connector-db-data:/var/lib/postgresql/data"
|
||||
restart: always
|
||||
|
||||
id-manager:
|
||||
image: docker.verbis.dkfz.de/ccp/idmanager:bridgehead-develop
|
||||
environment:
|
||||
MAGICPL_SITE: ${SITE}
|
||||
MAGICPL_MAINZELLISTE_API_KEY: ${MAGICPL_MAINZELLISTE_API_KEY}
|
||||
MAGICPL_API_KEY: ${MAGICPL_API_KEY}
|
||||
MAGICPL_API_KEY_CONNECTOR: ${MAGICPL_API_KEY_CONNECTOR}
|
||||
MAGICPL_MAINZELLISTE_CENTRAL_API_KEY: ${MAGICPL_MAINZELLISTE_CENTRAL_API_KEY}
|
||||
MAGICPL_CENTRAL_API_KEY: ${MAGICPL_CENTRAL_API_KEY}
|
||||
MAGICPL_OIDC_CLIENT_ID: ${MAGICPL_OIDC_CLIENT_ID}
|
||||
MAGICPL_OIDC_CLIENT_SECRET: ${MAGICPL_OIDC_CLIENT_SECRET}
|
||||
TOMCAT_REVERSEPROXY_FQDN: "${HOST}"
|
||||
HTTP_PROXY_USER: ${HTTP_PROXY_USER}
|
||||
HTTP_PROXY_PASSWORD: ${HTTP_PROXY_PASSWORD}
|
||||
HTTPS_PROXY_USER: ${HTTPS_PROXY_USER}
|
||||
HTTPS_PROXY_PASSWORD: ${HTTPS_PROXY_PASSWORD}
|
||||
env_file:
|
||||
- ../site-config/dktk.env
|
||||
volumes:
|
||||
- "id-manager-logs:/usr/local/tomcat/logs"
|
||||
depends_on:
|
||||
- patientlist
|
||||
labels:
|
||||
- "traefik.http.routers.id-manager.rule=PathPrefix(`/ID-Manager`)"
|
||||
- "traefik.http.services.id-manager.loadbalancer.server.port=8080"
|
||||
|
||||
patientlist:
|
||||
image: docker.verbis.dkfz.de/ccp/patientlist:bridgehead-develop
|
||||
environment:
|
||||
ML_SITE: ${SITE}
|
||||
ML_API_KEY: ${MAGICPL_MAINZELLISTE_API_KEY}
|
||||
ML_DB_PASS: ${ML_DB_PASS}
|
||||
TOMCAT_REVERSEPROXY_FQDN: "${HOST}"
|
||||
env_file:
|
||||
- ../site-config/dktk.env
|
||||
# TODO: Implement automatic seed generation in mainzelliste
|
||||
- ../site-config/patientlist.env
|
||||
volumes:
|
||||
- "patientlist-logs:/usr/local/tomcat/logs"
|
||||
labels:
|
||||
- "traefik.http.routers.patientlist.rule=PathPrefix(`/Patientlist`)"
|
||||
- "traefik.http.services.patientlist.loadbalancer.server.port=8080"
|
||||
depends_on:
|
||||
- patientlist-db
|
||||
|
||||
patientlist-db:
|
||||
image: postgres:13.1-alpine
|
||||
environment:
|
||||
POSTGRES_DB: mainzelliste
|
||||
POSTGRES_USER: mainzelliste
|
||||
POSTGRES_PASSWORD: ${ML_DB_PASS}
|
||||
TZ: "Europe/Berlin"
|
||||
volumes:
|
||||
- "patientlist-db-data:/var/lib/postgresql/data"
|
||||
@@ -1,69 +0,0 @@
|
||||
SITE=bridgehead_dktk_test
|
||||
SITEID=BRIDGEHEAD_DKTK_TEST
|
||||
|
||||
CONNECTOR_SHARE_URL="http://${HOST}:8080"
|
||||
CONNECTOR_ENABLE_METRICS=false
|
||||
CONNECTOR_MONITOR_INTERVAL=
|
||||
CONNECTOR_UPDATE_SERVER=
|
||||
|
||||
DEPLOYMENT_CONTEXT=dktk-connector
|
||||
|
||||
POSTGRES_PORT=5432
|
||||
POSTGRES_DB=share_v2
|
||||
POSTGRES_USER=samplyweb
|
||||
|
||||
HTTP_PROXY_HOST=${PROXY_URL}
|
||||
HTTP_PROXY_USER=
|
||||
HTTP_PROXY_PASSWORD=
|
||||
HTTPS_PROXY_HOST=${PROXY_URL}
|
||||
HTTPS_PROXY_USER=
|
||||
HTTPS_PROXY_PASSWORD=
|
||||
HTTP_PROXY=${PROXY_URL}
|
||||
HTTPS_PROXY=${PROXY_URL}
|
||||
http_proxy=${PROXY_URL}
|
||||
https_proxy=${PROXY_URL}
|
||||
|
||||
CCP_CENTRALSEARCH_URL=https://centralsearch-test.dktk.dkfz.de/
|
||||
CCP_DECENTRALSEARCH_URL=https://decentralsearch-test.ccp-it.dktk.dkfz.de/
|
||||
|
||||
CCP_MDR_URL=https://mdr.ccp-it.dktk.dkfz.de/v3/api/mdr
|
||||
MDR_URL=https://mdr.ccp-it.dktk.dkfz.de/v3/api/mdr
|
||||
|
||||
CCP_MONITOR_URL=
|
||||
MONITOR_OPTOUT=
|
||||
|
||||
PATIENTLIST_URL=http://bridgehead_patientlist:8080/Patientlist
|
||||
PROJECTPSEUDONYMISATION_URL=http://bridgehead_patientlist:8080/Patientlist
|
||||
|
||||
## nNGM
|
||||
#NNGM_URL=http://bridgeheadstore:8080
|
||||
#NNGM_PROFILE=http://uk-koeln.de/fhir/StructureDefinition/Patient/nNGM/pseudonymisiert
|
||||
#NNGM_MAINZELLISTE_URL=https://test.verbis.dkfz.de/mpl
|
||||
|
||||
##MDR
|
||||
MDR_NAMESPACE=adt,dktk,marker
|
||||
#MDR_MAP=
|
||||
MDR_VALIDATION=false
|
||||
|
||||
ML_DB_HOST=bridgehead_dktk_patientlist_db
|
||||
ML_DB_NAME=mainzelliste
|
||||
ML_DB_PORT=5432
|
||||
ML_DB_USER=mainzelliste
|
||||
|
||||
CENTRAL_CONTROL_NUMBER_GENERATPR_URL=http://e260-serv-03/central/api
|
||||
GLOBAL_ID=DKTK
|
||||
MAINZELLISTE_URL=https://patientlist-test.ccpit.dktk.dkfz.de/mainzelliste
|
||||
ML_DB_DRIVER=org.postgresql.Driver
|
||||
ML_DB_TYPE=postgresql
|
||||
|
||||
MAGICPL_MAINZELLISTE_URL=http://bridgehead_patientlist:8080
|
||||
|
||||
ML_LOG_LEVEL=warning
|
||||
ML_SITE=BRIDGEHEAD_DKTK_TEST
|
||||
TZ=Europe/Berlin
|
||||
|
||||
MAGICPL_SITE=adt
|
||||
MAGICPL_LOG_LEVEL=info
|
||||
MAGICPL_MAINZELLISTE_CENTRAL_URL=http://e260-serv-03/central/mainzelliste
|
||||
MAGICPL_CENTRAL_URL=http://e260-serv-03/central/api
|
||||
MAGICPL_OIDC_PROVIDER=https://auth-test.ccp-it.dktk.dkfz.de
|
||||
334
gbn/README.md
334
gbn/README.md
@@ -1,334 +0,0 @@
|
||||
# Bridgehead Deployment
|
||||
|
||||
|
||||
## Goal
|
||||
Allow the Sample Locator to search for patients and samples in your biobanks, giving researchers easy access to your resources.
|
||||
|
||||
|
||||
## Quick start
|
||||
If you simply want to set up a test installation, without exploring all of the possibilities offered by the Bridgehead, then the sections you need to look at are:
|
||||
* [Starting a Bridgehead](#starting-a-bridgehead)
|
||||
* [Register with a Sample Locator](#register-with-a-sample-locator)
|
||||
* [Checking your newly installed Bridgehead](#checking-your-newly-installed-bridgehead)
|
||||
|
||||
|
||||
## Background
|
||||
The **Sample Locator** is a tool that allows researchers to make searches for samples over a large number of geographically distributed biobanks. Each biobank runs a so-called **Bridgehead** at its site, which makes it visible to the Sample Locator. The Bridgehead is designed to give a high degree of protection to patient data. Additionally, a tool called the [Negotiator][negotiator] puts you in complete control over which samples and which data are delivered to which researcher.
|
||||
|
||||
You will most likely want to make your biobanks visible via the [publicly accessible Sample Locator][sl], but the possibility also exists to install your own Sample Locator for your site or organization, see the GitHub pages for [the server][sl-server-src] and [the GUI][sl-ui-src].
|
||||
|
||||
The Bridgehead has two primary components:
|
||||
* The **Blaze Store**. This is a highly responsive FHIR data store, which you will need to fill with your data via an ETL chain.
|
||||
* The **Connector**. This is the communication portal to the Sample Locator, with specially designed features that make it possible to run it behind a corporate firewall without making any compromises on security.
|
||||
|
||||
This document will show you how to:
|
||||
* Install the components making up the Bridgehead.
|
||||
* Register your Bridgehead with the Sample Locator, so that researchers can start searching your resources.
|
||||
|
||||
|
||||
## Requirements
|
||||
For data protection concept, server requirements, validation or import instructions, see [the list of general requirements][requirements].
|
||||
|
||||
|
||||
## Starting a Bridgehead
|
||||
The file `docker-compose.yml` contains the the minimum settings needed for installing and starting a Bridgehead on your computer. This Bridgehead should run straight out of the box. However, you may wish to modify this file, e.g. in order to:
|
||||
* Enable a corporate proxy (see below).
|
||||
* Set an alternative Sample Locator URL.
|
||||
* Change the admin credentials for the Connector.
|
||||
|
||||
To start a Bridgehead on your computer, you will need to follow the following steps:
|
||||
|
||||
* [Install Docker][docker] and [git][git]and test with:
|
||||
|
||||
```sh
|
||||
docker run hello-world
|
||||
git --version
|
||||
```
|
||||
|
||||
* Download this repository:
|
||||
|
||||
```sh
|
||||
git clone https://github.com/samply/bridgehead-deployment
|
||||
cd bridgehead-deployment
|
||||
```
|
||||
|
||||
* Launch the Bridgehead with the following command:
|
||||
|
||||
```sh
|
||||
docker-compose up -d
|
||||
```
|
||||
|
||||
* First test of the installation: check to see if there is a Connector running on port 8082:
|
||||
|
||||
```sh
|
||||
curl localhost:8082 | grep Welcome
|
||||
```
|
||||
|
||||
* If you need to stop the Bridgehead, from within this directory:
|
||||
|
||||
```sh
|
||||
docker-compose down
|
||||
```
|
||||
|
||||
## Port usage
|
||||
Once you have started the Bridgehead, the following components will be visible to you via ports on localhost:
|
||||
* Blaze Store: port 8080
|
||||
* Connector admin: port 8082
|
||||
|
||||
## Connector Administration
|
||||
The Connector administration panel allows you to set many of the parameters regulating your Bridgehead. Most especially, it is the place where you can register your site with the Sample Locator. To access this page, proceed as follows:
|
||||
|
||||
* Open the Connector page: http://localhost:8082
|
||||
* In the "Local components" box, click the "Samply Share" button.
|
||||
* A new page will be opened, where you will need to log in using the administrator credentials (admin/adminpass by default).
|
||||
* After log in, you will be taken to the administration dashboard, allowing you to configure the Connector.
|
||||
* If this is the first time you have logged in as an administrator, you are strongly recommended to set a more secure password! You can use the "Users" button on the dashboard to do this.
|
||||
|
||||
Note: your browser must be running on the same machine as the Connector for "localhost" URLs to work.
|
||||
|
||||
### Register with a Directory
|
||||
The [Directory][directory] is a BBMRI project that aims to catalog all biobanks in Europe and beyond. Each biobank is given its own unique ID and the Directory maintains counts of the number of donors and the number of samples held at each biobank. You are strongly encouraged to register with the Directory, because this opens the door to further services, such as the [Negotiator][negotiator].
|
||||
|
||||
Generally, you should register with the BBMRI national node for the country where your biobank is based. You can find a list of contacts for the national nodes [here](http://www.bbmri-eric.eu/national-nodes/). If your country is not in this list, or you have any questions, please contact the [BBMRI helpdesk](mailto:directory@helpdesk.bbmri-eric.eu). If your biobank is for COVID samples, you can also take advantage of an accelerated registration process [here](https://docs.google.com/forms/d/e/1FAIpQLSdIFfxADikGUf1GA0M16J0HQfc2NHJ55M_E47TXahju5BlFIQ).
|
||||
|
||||
Your national node will give you detailed instructions for registering, but for your information, here are the basic steps:
|
||||
|
||||
* Log in to the Directory for your country.
|
||||
* Add your biobank and enter its details, including contact information for a person involved in running the biobank.
|
||||
* You will need to create at least one collection.
|
||||
* Note the biobank ID and the collection ID that you have created - these will be needed when you register with the Locator (see below).
|
||||
|
||||
### Register with a Locator
|
||||
* Go to the registration page http://localhost:8082/admin/broker_list.xhtml.
|
||||
* To register with a Locator, enter the following values in the three fields under "Join new Searchbroker":
|
||||
* "Address": Depends on which Locator you want to register with:
|
||||
* `https://locator.bbmri-eric.eu/broker/`: BBMRI Locator production service (European).
|
||||
* `http://147.251.124.125:8088/broker/`: BBMRI Locator test service (European).
|
||||
* `https://samplelocator.bbmri.de/broker/`: GBA Sample Locator production service (German).
|
||||
* `https://samplelocator.test.bbmri.de/broker/`: GBA Sample Locator test service (German).
|
||||
* "Your email address": this is the email to which the registration token will be returned.
|
||||
* "Automatic reply": Set this to be `Total Size`
|
||||
* Click "Join" to start the registration process.
|
||||
* You should now have a list containing exactly one broker. You will notice that the "Status" box is empty.
|
||||
* Send an email to `feedback@germanbiobanknode.de` and let us know which of our Sample Locators you would like to register to. Please include the biobank ID and the collection ID from your Directory registration, if you have these available.
|
||||
* We will send you a registration token per email.
|
||||
* You will then re-open the Connector and enter the token into the "Status" box.
|
||||
* You should send us an email to let us know that you have done this.
|
||||
* We will then complete the registration process
|
||||
* We will email you to let you know that your biobank is now visible in the Sample Locator.
|
||||
|
||||
If you are a Sample Locator administrator, you will need to understand the [registration process](./SampleLocatorRegistration.md). Normal bridgehead admins do not need to worry about this.
|
||||
|
||||
### Monitoring
|
||||
You are strongly encouraged to set up an automated monitoring of your new Bridgehead. This will periodically test the Bridgehead in various ways, and (if you wish) will also send you emails if problems are detected. It helps you to become aware of problems before users do, and also gives you the information you need to track down the source of the problems. To activate monitoring, perform the following steps:
|
||||
|
||||
* Open the Connector administration dashboard in your browser, see [Admin](#connector-administration) for details.
|
||||
* Click the "Configuration" button.
|
||||
* 
|
||||
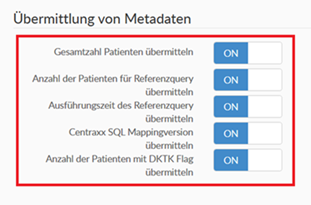
* Scroll to the section "Reporting to central services".
|
||||
* Click on all of the services in this section so that they have the status "ON".
|
||||
* 
|
||||
* **Important:** Scroll to the bottom of the page and click the "Save" button.
|
||||
* 
|
||||
* Return to the dashboard, and click the button "Scheduled Tasks".
|
||||
* 
|
||||
* Scroll down to the box labelled "ReportToMonitoringJob". For newer Versions of the bridgehead there this job is separated into "ReportToMonitoringJobShortFrequence" and "ReportToMonitoringJobLongFrequence".
|
||||
* Click the button "Run now". This switches the monitoring on. If you have the newer Version of the bridgeheads please run both jobs.
|
||||
* 
|
||||
* If you want to receive emails when the monitoring service detects problems with your Bridgehead, please send a list of email addresses for the people to be notified to: `feedback@germanbiobanknode.de`.
|
||||
|
||||
You are now done!
|
||||
|
||||
### Troubleshooting
|
||||
To get detailled information about Connector problems, you need to use the Docker logging facility:
|
||||
|
||||
* Log into the server where the Connector is running. You need a command line login.
|
||||
* Discover the container ID of the Connector. First run "docker ps". Look in the list of results. The relevant line contains the image name "samply/share-client".
|
||||
* Execute the following command: "docker logs \<Container-ID\>"
|
||||
* The last 100 lines of the log are relevant. Maybe you will see the problem there right away. Otherwise, send the log-selection to us.
|
||||
|
||||
### User
|
||||
* To enable a user to access the connector, a new user can be created under http://localhost:8082/admin/user_list.xhtml.
|
||||
This user has the possibility to view incoming queries
|
||||
|
||||
### Jobs
|
||||
* The connector uses [Quartz Jobs](http://www.quartz-scheduler.org/) to do things like collect the queries from the searchbroker or execute the queries.
|
||||
The full list of jobs can be viewed under the job page http://localhost:8082/admin/job_list.xhtml.
|
||||
|
||||
### Tests
|
||||
* The Connector connectivity checks can be found under the test page http://localhost:8082/admin/tests.xhtml.
|
||||
|
||||
## Checking your newly installed Bridgehead
|
||||
We will load some test data and then run a query to see if it shows up.
|
||||
|
||||
First, install [bbmri-fhir-gen][bbmri-fhir-gen]. Run the following command:
|
||||
|
||||
```sh
|
||||
mkdir TestData
|
||||
bbmri-fhir-gen TestData -n 10
|
||||
```
|
||||
|
||||
This will generate test data for 10 patients, and place it in the directory `TestData`.
|
||||
|
||||
Next, install [blazectl][blazectl]. Run the following commands:
|
||||
|
||||
```sh
|
||||
blazectl --server http://localhost:8080/fhir upload TestData
|
||||
blazectl --server http://localhost:8080/fhir count-resources
|
||||
```
|
||||
|
||||
If both of them complete successfully, it means that the test data has successfully been uploaded to the Blaze Store.
|
||||
|
||||
Open the [Sample Locator][sl] and hit the "SEND" button. You may need to wait for a minute before all results are returned. Click the "Login" link to log in via the academic institution where you work (AAI). You should now see a list of the biobanks known to the Sample Locator.
|
||||
|
||||
If your biobank is present, and it contains non-zero counts of patients and samples, then your installation was successful.
|
||||
|
||||
If you wish to remove the test data, you can do so by simply deleting the Docker volume for the Blaze Store database:
|
||||
|
||||
```sh
|
||||
docker-compose down
|
||||
docker volume rm store-db-data
|
||||
```
|
||||
|
||||
## Manual installation
|
||||
The installation described here uses Docker, meaning that you don't have to worry about configuring or installing the Bridgehead components - Docker does it all for you. If you do not wish to use Docker, you can install all of the software directly on your machine, as follows:
|
||||
|
||||
* Install the [Blaze Store][man-store]
|
||||
* Install the [Connector][man-connector]
|
||||
* Register with the Sample Locator (see above)
|
||||
|
||||
|
||||
Source code for components deployed by `docker-compose.yml`:
|
||||
|
||||
* [Store][store-src]
|
||||
* [Connector][connector-src]
|
||||
|
||||
|
||||
## Optional configuration:
|
||||
|
||||
#### Proxy example
|
||||
Add environments variables in `docker-compose.yml` (remove user and password environments if not available):
|
||||
"http://proxy.example.de:8080", user "testUser", password "testPassword"
|
||||
|
||||
version: '3.4'
|
||||
services:
|
||||
store:
|
||||
container_name: "store"
|
||||
image: "samply/blaze:0.11"
|
||||
environment:
|
||||
BASE_URL: "http://store:8080"
|
||||
JAVA_TOOL_OPTIONS: "-Xmx4g"
|
||||
PROXY_HOST: "http://proxy.example.de"
|
||||
PROXY_PORT: "8080"
|
||||
PROXY_USER: "testUser"
|
||||
PROXY_PASSWORD: "testPassword"
|
||||
networks:
|
||||
- "samply"
|
||||
|
||||
.......
|
||||
|
||||
connector:
|
||||
container_name: "connector"
|
||||
image: "samply/connector:7.0.0"
|
||||
environment:
|
||||
POSTGRES_HOST: "connector-db"
|
||||
POSTGRES_DB: "samply.connector"
|
||||
POSTGRES_USER: "samply"
|
||||
POSTGRES_PASS: "samply"
|
||||
STORE_URL: "http://store:8080/fhir"
|
||||
QUERY_LANGUAGE: "CQL"
|
||||
MDR_URL: "https://mdr.germanbiobanknode.de/v3/api/mdr"
|
||||
HTTP_PROXY: "http://proxy.example.de:8080"
|
||||
PROXY_USER: "testUser"
|
||||
PROXY_PASS: "testPassword
|
||||
networks:
|
||||
- "samply"
|
||||
|
||||
.......
|
||||
|
||||
|
||||
|
||||
#### General information on Docker environment variables used in the Bridgehead
|
||||
|
||||
* [Store][env-store]
|
||||
* [Connector][env-connector]
|
||||
|
||||
|
||||
## Notes
|
||||
* If you see database connection errors of Store or Connector, open a second terminal and run `docker-compose stop` followed by `docker-compose start`. Database connection problems should only occur at the first start because the store and the connector doesn't wait for the databases to be ready. Both try to connect at startup which might be to early.
|
||||
|
||||
* If one needs only one of the the Bridgehead components, they can be started individually:
|
||||
|
||||
```sh
|
||||
docker-compose up store -d
|
||||
docker-compose up connector -d
|
||||
```
|
||||
|
||||
* To shut down all services (but keep databases):
|
||||
|
||||
```sh
|
||||
docker-compose down
|
||||
```
|
||||
|
||||
* To delete databases as well (destroy before):
|
||||
|
||||
```sh
|
||||
docker volume rm store-db-data
|
||||
docker volume rm connector-db-data
|
||||
```
|
||||
|
||||
* To see all executed queries, create a [new user][connector-user], logout and login with this normal user.
|
||||
|
||||
* To set Store-Basic-Auth-credentials in Connector (as default `Lokales Datenmanagement` with dummy values was generated)
|
||||
* Login at [Connector-UI][connector-login] (default usr=admin, pwd=adminpass)
|
||||
* Open [credentials page][connector-credentials]
|
||||
- Delete all instances of`Lokales Datenmanagement`
|
||||
- for "Ziel" select `Lokales Datenmanagement`, provide decrypted CREDENTIALS in "Benutzername" and "Passwort", select "Zugangsdaten hinzufügen"
|
||||
|
||||
* If you would like to read about the experiences of a team in Brno who have installed the Bridgehead and a local Sample Locator instance, take a look at [SL-BH_deploy](SL-BH_deploy).
|
||||
|
||||
## Useful Links
|
||||
* [FHIR Quality Reporting Authoring UI][quality-ui-github]
|
||||
* [How to join Sample Locator][join-sl]
|
||||
* [Samply code repositories][samply]
|
||||
|
||||
## License
|
||||
|
||||
Copyright 2019 - 2021 The Samply Community
|
||||
|
||||
Licensed under the Apache License, Version 2.0 (the "License"); you may not use this file except in compliance with the License. You may obtain a copy of the License at
|
||||
|
||||
http://www.apache.org/licenses/LICENSE-2.0
|
||||
|
||||
Unless required by applicable law or agreed to in writing, software distributed under the License is distributed on an "AS IS" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. See the License for the specific language governing permissions and limitations under the License.
|
||||
|
||||
[sl]: <https://samplelocator.bbmri.de>
|
||||
[sl-ui-src]: <https://github.com/samply/sample-locator>
|
||||
[sl-server-src]: <https://github.com/samply/share-broker-rest>
|
||||
[negotiator]: <https://negotiator.bbmri-eric.eu/login.xhtml>
|
||||
[directory]: <https://directory.bbmri-eric.eu>
|
||||
[bbmri]: <http://www.bbmri-eric.eu>
|
||||
[docker]: <https://docs.docker.com/install>
|
||||
[git]: <https://www.atlassian.com/git/tutorials/install-git>
|
||||
|
||||
[connector-user]:<http://localhost:8082/admin/user_list.xhtml>
|
||||
[connector-login]:<http://localhost:8082/admin/login.xhtml>
|
||||
[connector-credentials]:<http://localhost:8082/admin/credentials_list.xhtml>
|
||||
|
||||
[requirements]: <https://samply.github.io/bbmri-fhir-ig/howtoJoin.html#general-requirements>
|
||||
|
||||
[man-store]: <https://github.com/samply/blaze/blob/master/docs/deployment/manual-deployment.md>
|
||||
[env-store]: <https://github.com/samply/blaze/blob/master/docs/deployment/environment-variables.md>
|
||||
[env-connector]: <https://github.com/samply/share-client/blob/master/docs/deployment/docker-deployment.md>
|
||||
|
||||
[bbmri-fhir-gen]: <https://github.com/samply/bbmri-fhir-gen>
|
||||
[blazectl]: <https://github.com/samply/blazectl>
|
||||
|
||||
[man-connector]: <Connector.md>
|
||||
|
||||
[store-src]: <https://github.com/samply/blaze>
|
||||
[connector-src]: <https://github.com/samply/share-client>
|
||||
|
||||
[quality-ui-github]:<https://github.com/samply/blaze-quality-reporting-ui>
|
||||
[join-sl]: <https://samply.github.io/bbmri-fhir-ig/howtoJoin.html>
|
||||
[samply]: <https://github.com/samply>
|
||||
@@ -53,9 +53,6 @@ services:
|
||||
depends_on:
|
||||
- "gbn-connector-db"
|
||||
restart: "always"
|
||||
env_file:
|
||||
###TODO: Move into Image
|
||||
- internal.env
|
||||
|
||||
gbn-connector-db:
|
||||
image: "postgres:10.17"
|
||||
|
||||
@@ -1,16 +0,0 @@
|
||||
POSTGRES_DB=samply.connector
|
||||
POSTGRES_USER=samply
|
||||
QUERY_LANGUAGE=CQL
|
||||
CONNECTOR_SHARE_URL="http://${HOST}:8080"
|
||||
MDR_URL="https://mdr.germanbiobanknode.de/v3/api/mdr"
|
||||
MONITOR_URL=https://ccpit.dktk.dkfz.de/dktk.monitor/rest/info
|
||||
DEPLOYMENT_CONTEXT=gbn-connector
|
||||
CONNECTOR_MONITOR_INTERVAL=
|
||||
CONNECTOR_UPDATE_SERVER=
|
||||
feature_BBMRI_DIRECTORY_SYNC=false
|
||||
feature_DKTK_CENTRAL_SEARCH=false
|
||||
feature_NNGM_CTS=false
|
||||
LOG_LEVEL=info
|
||||
STORE_URL=http://bridgehead_gbn_blaze_store:8080
|
||||
|
||||
CONNECTOR_ENABLE_METRICS=false
|
||||
@@ -1,444 +0,0 @@
|
||||
<?xml version="1.0" encoding="UTF-8"?>
|
||||
<!DOCTYPE pathconfig [
|
||||
<!ENTITY localPatientListUrl "MAGICPL_MAINZELLISTE_URL">
|
||||
<!ENTITY localPatientlistApiKey "MAGICPL_MAINZELLISTE_API_KEY">
|
||||
<!ENTITY magicplApiKey "MAGICPL_API_KEY">
|
||||
<!ENTITY magicplApiKeyConnector "MAGICPL_API_KEY_CONNECTOR">
|
||||
<!ENTITY centralPatientListUrl "MAGICPL_MAINZELLISTE_CENTRAL_URL">
|
||||
<!ENTITY centralPatientListApiKey "MAGICPL_MAINZELLISTE_CENTRAL_API_KEY">
|
||||
<!ENTITY centralKNEUrl "MAGICPL_CENTRAL_URL">
|
||||
<!ENTITY centralKNEApiKey "MAGICPL_CENTRAL_API_KEY">
|
||||
<!ENTITY site "MAGICPL_SITE">
|
||||
]>
|
||||
<pathconfig xmlns="http://www.example.org/Pfade" xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance"
|
||||
xsi:schemaLocation="http://www.example.org/Pfade magicpl.xsd ">
|
||||
<paths>
|
||||
<multipath>
|
||||
<name>getId</name>
|
||||
<input>
|
||||
<iorecord ref="IDAT" name="IDAT"/>
|
||||
<iosingle ref="extIDs" name="extIDs"/>
|
||||
<iosingle ref="idTypes" name="idTypes"/>
|
||||
<iosingle ref="consented" name="consented"/>
|
||||
<iosingle ref="auditTrail" name="auditTrail"/>
|
||||
<iosingle ref="sureness" name="sureness"/>
|
||||
</input>
|
||||
<output>
|
||||
<iosingle ref="ids" name="ids"/>
|
||||
</output>
|
||||
<step>
|
||||
<name>getRequestedId</name>
|
||||
<parameters/>
|
||||
<input>
|
||||
<iorecord ref="IDAT" name="IDAT"/>
|
||||
<iosingle ref="idTypes" name="idTypes"/>
|
||||
<iosingle ref="consented" name="consented"/>
|
||||
<iosingle ref="auditTrail" name="auditTrail"/>
|
||||
<iosingle ref="sureness" name="sureness"/>
|
||||
</input>
|
||||
<output>
|
||||
<iosingle ref="ids" name="ids"/>
|
||||
</output>
|
||||
<switch>
|
||||
<evaluator name="de.pseudonymisierung.magicpl.evaluator.IsConsentedEvaluator">
|
||||
<parameters>
|
||||
<parameter name="key">consented</parameter>
|
||||
</parameters>
|
||||
</evaluator>
|
||||
<!-- eventuell muss hier andere unterscheidung genommen werden -->
|
||||
<case value="true">
|
||||
<multipath>
|
||||
<name>getDktkIdWithIdat</name>
|
||||
<parameters/>
|
||||
<input>
|
||||
<iorecord ref="IDAT"/>
|
||||
<iosingle ref="idTypes" name="idTypes"/>
|
||||
<iosingle ref="extIDs" name="extIDs"/>
|
||||
<iosingle ref="consented"/>
|
||||
<iosingle ref="auditTrail" name="auditTrail"/>
|
||||
<iosingle ref="sureness" name="sureness"/>
|
||||
</input>
|
||||
<output>
|
||||
<iosingle ref="ids" name="ids"/>
|
||||
</output>
|
||||
<step>
|
||||
<name>getIdToken</name>
|
||||
<parameters>
|
||||
<!-- Hier war lokale Pfade URL angegeben, es soll aber eigentlich zentraler KNE aufgerufen werden -->
|
||||
<parameter name="url">¢ralKNEUrl;/paths/getIdToken</parameter>
|
||||
<parameter name="method">POST</parameter>
|
||||
<parameter name="apiKey">¢ralKNEApiKey;</parameter>
|
||||
<parameter name="excludeIdTypes">BK_&site;_L-ID</parameter>
|
||||
<parameter name="excludeIDAT">locallyUniqueId</parameter>
|
||||
</parameters>
|
||||
<input>
|
||||
<iorecord ref="IDAT" name="IDAT"/>
|
||||
<iosingle ref="idTypes" name="idTypes"/>
|
||||
<iosingle ref="auditTrail" name="auditTrail"/>
|
||||
</input>
|
||||
<output>
|
||||
<iosingle ref="tokenId"/>
|
||||
</output>
|
||||
<implementation>de.pseudonymisierung.magicpl.processor.PathClient</implementation>
|
||||
</step>
|
||||
<step>
|
||||
<name>getDktkId</name>
|
||||
<parameters>
|
||||
<parameter name="mainzellisteURL">¢ralPatientListUrl;</parameter>
|
||||
<parameter name="mainzellisteApiKey">¢ralPatientListApiKey;</parameter>
|
||||
<parameter name="mainzellisteApiVersion">3.2</parameter>
|
||||
</parameters>
|
||||
<input>
|
||||
<iosingle ref="tokenId"/>
|
||||
<iosingle ref="sureness" name="sureness"/>
|
||||
</input>
|
||||
<output>
|
||||
<iosingle ref="ids" name="ids"/>
|
||||
</output>
|
||||
<implementation>de.pseudonymisierung.magicpl.processor.MainzellisteClient</implementation>
|
||||
</step>
|
||||
<step>
|
||||
<name>mapIdStringToExternalId</name>
|
||||
<parameters>
|
||||
<parameter name="map">ids->extIDs</parameter>
|
||||
</parameters>
|
||||
<input>
|
||||
<iosingle ref="ids" name="ids"/>
|
||||
</input>
|
||||
<output>
|
||||
<iosingle ref="extIDs" name="extIDs"/>
|
||||
</output>
|
||||
<implementation>de.pseudonymisierung.magicpl.processor.InputMapperProcessor</implementation>
|
||||
</step>
|
||||
<step>
|
||||
<name>getLocalTKTExternalId</name>
|
||||
<parameters>
|
||||
<parameter name="mainzellisteURL">&localPatientListUrl;</parameter>
|
||||
<parameter name="mainzellisteApiKey">&localPatientlistApiKey;</parameter>
|
||||
<parameter name="fieldType">plain</parameter>
|
||||
</parameters>
|
||||
<input>
|
||||
<iorecord ref="IDAT" name="IDAT"/>
|
||||
<iosingle ref="extIDs" name="extIDs"/>
|
||||
<iosingle ref="idTypes" name="idTypes"/>
|
||||
<iosingle ref="auditTrail" name="auditTrail"/>
|
||||
</input>
|
||||
<output>
|
||||
<iosingle ref="tokenId"/>
|
||||
</output>
|
||||
<!-- TODO: Implementation für Ticket Erzeugung ohne Kontrolnummer -->
|
||||
<!-- Wird hier eine neue Klasse benötigt oder kann diese hier entsprechend angepasst werden? -->
|
||||
<implementation>de.pseudonymisierung.magicpl.processor.MainzellisteKNTicketClient</implementation>
|
||||
</step>
|
||||
<step>
|
||||
<name>generateLocalIdExternalId</name>
|
||||
<parameters>
|
||||
<parameter name="mainzellisteURL">&localPatientListUrl;</parameter>
|
||||
<parameter name="mainzellisteApiKey">&localPatientlistApiKey;</parameter>
|
||||
<parameter name="mainzellisteApiVersion">3.2</parameter>
|
||||
<parameter name="idType">BK_&site;_L-ID</parameter>
|
||||
</parameters>
|
||||
<input>
|
||||
<iosingle ref="tokenId"/>
|
||||
<iosingle ref="extIDs" name="extIDs"/>
|
||||
<iosingle ref="sureness" name="sureness"/>
|
||||
</input>
|
||||
<output>
|
||||
<iosingle ref="ids" name="ids"/>
|
||||
</output>
|
||||
<implementation>de.pseudonymisierung.magicpl.processor.MainzellisteClient</implementation>
|
||||
</step>
|
||||
</multipath>
|
||||
</case>
|
||||
<case value="false">
|
||||
<multipath>
|
||||
<name>getLocalIdWithIDAT</name>
|
||||
<input>
|
||||
<iorecord ref="IDAT" name="IDAT"/>
|
||||
<iosingle ref="idTypes" name="idTypes"/>
|
||||
<iosingle ref="auditTrail" name="auditTrail"/>
|
||||
<iosingle ref="sureness" name="sureness"/>
|
||||
</input>
|
||||
<output>
|
||||
<iosingle ref="ids" name="ids"/>
|
||||
</output>
|
||||
<step>
|
||||
<name>getTKT</name>
|
||||
<parameters>
|
||||
<parameter name="mainzellisteURL">&localPatientListUrl;</parameter>
|
||||
<parameter name="mainzellisteApiKey">&localPatientlistApiKey;</parameter>
|
||||
<parameter name="fieldType">plain</parameter>
|
||||
</parameters>
|
||||
<input>
|
||||
<iorecord ref="IDAT" name="IDAT"/>
|
||||
<iosingle ref="idTypes" name="idTypes"/>
|
||||
<iosingle ref="auditTrail" name="auditTrail"/>
|
||||
</input>
|
||||
<output>
|
||||
<iosingle ref="tokenId"/>
|
||||
</output>
|
||||
<!-- Wird hier eine neue Klasse benötigt oder kann diese hier entsprechend angepasst werden? -->
|
||||
<implementation>de.pseudonymisierung.magicpl.processor.MainzellisteKNTicketClient</implementation>
|
||||
</step>
|
||||
<step>
|
||||
<name>generateLocalId</name>
|
||||
<parameters>
|
||||
<parameter name="mainzellisteURL">&localPatientListUrl;</parameter>
|
||||
<parameter name="mainzellisteApiKey">&localPatientlistApiKey;</parameter>
|
||||
<parameter name="mainzellisteApiVersion">3.2</parameter>
|
||||
<parameter name="idType">BK_&site;_L-ID</parameter>
|
||||
</parameters>
|
||||
<input>
|
||||
<iosingle ref="tokenId"/>
|
||||
<iosingle ref="sureness" name="sureness"/>
|
||||
</input>
|
||||
<output>
|
||||
<iosingle ref="ids" name="ids"/>
|
||||
</output>
|
||||
<implementation>de.pseudonymisierung.magicpl.processor.MainzellisteClient</implementation>
|
||||
</step>
|
||||
</multipath>
|
||||
</case>
|
||||
</switch>
|
||||
</step>
|
||||
</multipath>
|
||||
<multipath>
|
||||
<name>readPatients</name>
|
||||
<input>
|
||||
<iosingle ref="searchIds" name="searchIds"/>
|
||||
<iosingle ref="resultIds" name="resultIds"/>
|
||||
<iosingle ref="auditTrail" name="auditTrail"/>
|
||||
</input>
|
||||
<output>
|
||||
<iosingle ref="patients" name="patients"/>
|
||||
</output>
|
||||
<step>
|
||||
<name>DecideLocalOrCentralRequest</name>
|
||||
<input>
|
||||
<iosingle ref="searchIds" name="searchIds"/>
|
||||
<iosingle ref="resultIds" name="resultIds"/>
|
||||
<iosingle ref="auditTrail" name="auditTrail"/>
|
||||
</input>
|
||||
<output>
|
||||
<iosingle ref="patients" name="patients"/>
|
||||
</output>
|
||||
<switch>
|
||||
<evaluator name="de.pseudonymisierung.magicpl.evaluator.RegexEvaluator">
|
||||
<parameters>
|
||||
<parameter name="regex">MDS_\*_G-ID</parameter>
|
||||
<parameter name="fieldName">resultIds</parameter>
|
||||
</parameters>
|
||||
</evaluator>
|
||||
<case value="true">
|
||||
<multipath>
|
||||
<name>readPatientsCentrally</name>
|
||||
<input>
|
||||
<iosingle ref="searchIds" name="searchIds"/>
|
||||
<iosingle ref="resultIds" name="resultIds"/>
|
||||
<iosingle ref="auditTrail" name="auditTrail"/>
|
||||
</input>
|
||||
<output>
|
||||
<iosingle ref="patients" name="patients"/>
|
||||
</output>
|
||||
<step>
|
||||
<name>createReadTokenCentral</name>
|
||||
<parameters>
|
||||
<parameter name="mainzellisteURL">¢ralPatientListUrl;</parameter>
|
||||
<parameter name="mainzellisteApiKey">¢ralPatientListApiKey;</parameter>
|
||||
<parameter name="ignoredIdTypeCheck">true</parameter>
|
||||
</parameters>
|
||||
<input>
|
||||
<iosingle ref="searchIds" name="searchIds"/>
|
||||
<iosingle ref="resultIds" name="resultIds"/>
|
||||
<iosingle ref="auditTrail" name="auditTrail"/>
|
||||
</input>
|
||||
<output>
|
||||
<iosingle ref="tokenId"/>
|
||||
</output>
|
||||
<implementation>de.pseudonymisierung.magicpl.processor.MainzellisteReadTokenClient</implementation>
|
||||
</step>
|
||||
<step>
|
||||
<name>resolveReadTokenCentral</name>
|
||||
<parameters>
|
||||
<parameter name="mainzellisteURL">¢ralPatientListUrl;</parameter>
|
||||
<parameter name="mainzellisteApiKey">¢ralPatientListApiKey;</parameter>
|
||||
</parameters>
|
||||
<input>
|
||||
<iosingle ref="tokenId"/>
|
||||
</input>
|
||||
<output>
|
||||
<iosingle ref="patients"/>
|
||||
</output>
|
||||
<implementation>de.pseudonymisierung.magicpl.processor.MainzellisteReadClient</implementation>
|
||||
</step>
|
||||
</multipath>
|
||||
</case>
|
||||
<case value="false">
|
||||
<multipath>
|
||||
<name>readPatientsLocally</name>
|
||||
<input>
|
||||
<iosingle ref="searchIds" name="searchIds"/>
|
||||
<iosingle ref="resultIds" name="resultIds"/>
|
||||
<iosingle ref="auditTrail" name="auditTrail"/>
|
||||
</input>
|
||||
<output>
|
||||
<iosingle ref="patients" name="patients"/>
|
||||
</output>
|
||||
<step>
|
||||
<name>createReadToken</name>
|
||||
<parameters>
|
||||
<parameter name="mainzellisteURL">&localPatientListUrl;</parameter>
|
||||
<parameter name="mainzellisteApiKey">&localPatientlistApiKey;</parameter>
|
||||
<parameter name="ignoredIdTypeCheck">true</parameter>
|
||||
</parameters>
|
||||
<input>
|
||||
<iosingle ref="searchIds" name="searchIds"/>
|
||||
<iosingle ref="resultIds" name="resultIds"/>
|
||||
<iosingle ref="auditTrail" name="auditTrail"/>
|
||||
</input>
|
||||
<output>
|
||||
<iosingle ref="tokenId"/>
|
||||
</output>
|
||||
<implementation>de.pseudonymisierung.magicpl.processor.MainzellisteReadTokenClient</implementation>
|
||||
</step>
|
||||
<step>
|
||||
<name>resolveReadToken</name>
|
||||
<parameters>
|
||||
<parameter name="mainzellisteURL">&localPatientListUrl;</parameter>
|
||||
<parameter name="mainzellisteApiKey">&localPatientlistApiKey;</parameter>
|
||||
</parameters>
|
||||
<input>
|
||||
<iosingle ref="tokenId"/>
|
||||
</input>
|
||||
<output>
|
||||
<iosingle ref="patients"/>
|
||||
</output>
|
||||
<implementation>de.pseudonymisierung.magicpl.processor.MainzellisteReadClient</implementation>
|
||||
</step>
|
||||
</multipath>
|
||||
</case>
|
||||
</switch>
|
||||
</step>
|
||||
</multipath>
|
||||
</paths>
|
||||
|
||||
<iodefinitions>
|
||||
<iorecord name="IDAT">
|
||||
<iosingle name="Vorname"/>
|
||||
<iosingle name="Nachname"/>
|
||||
<iosingle name="Fruehere_Namen"/>
|
||||
<iosingle name="Geburtstag"/>
|
||||
<iosingle name="Geburtsmonat"/>
|
||||
<iosingle name="Geburtsjahr"/>
|
||||
<iosingle name="Staatsangehoerigkeit"/>
|
||||
<iosingle name="Geschlecht"/>
|
||||
<!-- external id which will be inputed into paths -->
|
||||
<iosingle name="locallyUniqueId"/>
|
||||
</iorecord>
|
||||
<iosingle name="idTypes" type="ArrayList"/>
|
||||
<iosingle name="ids"/>
|
||||
<!-- iosingle for external Ids which will be transferred between two steps -->
|
||||
<iosingle name="extIDs"/>
|
||||
<iosingle name="consented"/>
|
||||
<iosingle name="tokenId"/>
|
||||
<iosingle name="auditTrail"/>
|
||||
<iosingle name="searchIds"/>
|
||||
<iosingle name="resultIds"/>
|
||||
<iosingle name="patients"/>
|
||||
<iosingle name="sureness"/>
|
||||
</iodefinitions>
|
||||
<authentication>
|
||||
<client>
|
||||
<permissions>getId</permissions>
|
||||
<restrictions>
|
||||
<restriction path="getId">
|
||||
<inputRestriction input="idTypes">
|
||||
<permissibleValues>
|
||||
<value>DKTK000001950_&site;_L-ID</value>
|
||||
<value>DKTK000001950_&site;_G-ID</value>
|
||||
</permissibleValues>
|
||||
</inputRestriction>
|
||||
</restriction>
|
||||
</restrictions>
|
||||
<roles>DKTK000001950_USER</roles>
|
||||
</client>
|
||||
<client>
|
||||
<permissions>getId</permissions>
|
||||
<restrictions>
|
||||
<restriction path="getId">
|
||||
<inputRestriction input="idTypes">
|
||||
<permissibleValues>
|
||||
<value>DKTK000001951_&site;_L-ID</value>
|
||||
<value>DKTK000001951_&site;_G-ID</value>
|
||||
</permissibleValues>
|
||||
</inputRestriction>
|
||||
</restriction>
|
||||
</restrictions>
|
||||
<roles>DKTK000001951_USER</roles>
|
||||
</client>
|
||||
<client>
|
||||
<permissions>getId</permissions>
|
||||
<restrictions>
|
||||
<restriction path="getId">
|
||||
<inputRestriction input="idTypes">
|
||||
<permissibleValues>
|
||||
<value>DKTK000001985_&site;_L-ID</value>
|
||||
<value>DKTK000001985_&site;_G-ID</value>
|
||||
</permissibleValues>
|
||||
</inputRestriction>
|
||||
</restriction>
|
||||
</restrictions>
|
||||
<roles>DKTK000001985_USER</roles>
|
||||
</client>
|
||||
<client>
|
||||
<permissions>getId</permissions>
|
||||
<restrictions>
|
||||
<restriction path="getId">
|
||||
<inputRestriction input="idTypes">
|
||||
<permissibleValues>
|
||||
<value>DKTK000001986_&site;_L-ID</value>
|
||||
<value>DKTK000001986_&site;_G-ID</value>
|
||||
</permissibleValues>
|
||||
</inputRestriction>
|
||||
</restriction>
|
||||
</restrictions>
|
||||
<roles>DKTK000001986_USER</roles>
|
||||
</client>
|
||||
<client>
|
||||
<permissions>getId</permissions>
|
||||
<restrictions>
|
||||
<restriction path="getId">
|
||||
<inputRestriction input="idTypes">
|
||||
<permissibleValues>
|
||||
<value>DKTK999999999_&site;_L-ID</value>
|
||||
<value>DKTK999999999_&site;_G-ID</value>
|
||||
</permissibleValues>
|
||||
</inputRestriction>
|
||||
</restriction>
|
||||
</restrictions>
|
||||
<roles>DKTK999999999_USER</roles>
|
||||
</client>
|
||||
<client>
|
||||
<permissions>getId</permissions>
|
||||
<restrictions>
|
||||
<restriction path="getId">
|
||||
<inputRestriction input="idTypes">
|
||||
<permissibleValues>
|
||||
<value>DKTK000002089_&site;_L-ID</value>
|
||||
<value>DKTK000002089_&site;_G-ID</value>
|
||||
</permissibleValues>
|
||||
</inputRestriction>
|
||||
</restriction>
|
||||
</restrictions>
|
||||
<roles>DKTK000002089_USER</roles>
|
||||
</client>
|
||||
<client>
|
||||
<permissions>getId</permissions>
|
||||
<apiKey>&magicplApiKey;</apiKey>
|
||||
</client>
|
||||
<client>
|
||||
<permissions>readPatients</permissions>
|
||||
<apiKey>&magicplApiKeyConnector;</apiKey>
|
||||
</client>
|
||||
</authentication>
|
||||
</pathconfig>
|
||||
@@ -1,37 +0,0 @@
|
||||
# Konfiguration for the bridgehead
|
||||
server {
|
||||
|
||||
# this is the internal Docker DNS, cache only for 30s
|
||||
resolver 127.0.0.11 valid=30s;
|
||||
|
||||
listen 80;
|
||||
listen [::]:80;
|
||||
|
||||
proxy_set_header Host $host;
|
||||
proxy_set_header X-Real-IP $remote_addr;
|
||||
proxy_set_header X-Forwarded-For $proxy_add_x_forwarded_for;
|
||||
proxy_set_header X-Forwarded-Host $proxy_host;
|
||||
proxy_set_header X-Forwarded-Proto $scheme;
|
||||
|
||||
### Defining the urls like this, will stop nginx from failing on startup if one service isn't started.
|
||||
set $internal_connector_url http://connector:8080;
|
||||
set $internal_idmanager_url http://idmanager:8080;
|
||||
set $internal_patientlist_url http://patientlist:8080;
|
||||
set $internal_ldm_url http://ldm:8080;
|
||||
|
||||
location / {
|
||||
proxy_pass $internal_connector_url;
|
||||
}
|
||||
|
||||
location /ID-Manager/ {
|
||||
proxy_pass $internal_idmanager_url;
|
||||
}
|
||||
|
||||
location /Patientlist/ {
|
||||
proxy_pass $internal_patientlist_url;
|
||||
}
|
||||
|
||||
location /LocalDatamanagement/ {
|
||||
proxy_pass $internal_ldm_url;
|
||||
}
|
||||
}
|
||||
29
lib/add_bc_user.sh
Executable file
29
lib/add_bc_user.sh
Executable file
@@ -0,0 +1,29 @@
|
||||
#!/bin/bash -e
|
||||
|
||||
echo "This script add's a user with password to the bridghead"
|
||||
|
||||
if [ $# -eq 0 ]; then
|
||||
echo "No arguments provided, please provide the project name"
|
||||
exit 1
|
||||
fi
|
||||
|
||||
if [ ! -f /etc/systemd/system/bridgehead@$1.service.d/override.conf ]; then
|
||||
echo "Please create a Service first, with setup-bridgehead-units.sh"
|
||||
exit
|
||||
fi
|
||||
|
||||
read -p 'Username: ' bc_user
|
||||
read -sp 'Password: ' bc_password
|
||||
|
||||
echo
|
||||
|
||||
|
||||
bc=`docker run --rm -it httpd:latest htpasswd -nb $bc_user $bc_password`
|
||||
|
||||
if grep -q -E "Environment=bc_auth_users=" /etc/systemd/system/bridgehead@$1.service.d/override.conf ; then
|
||||
x=`grep -E "Environment=bc_auth_users=" /etc/systemd/system/bridgehead@$1.service.d/override.conf`
|
||||
sed -i "/Environment=bc_auth_users=/c\\$x,$bc" /etc/systemd/system/bridgehead@$1.service.d/override.conf
|
||||
|
||||
else
|
||||
echo "Environment=bc_auth_users=${bc}" >> /etc/systemd/system/bridgehead@$1.service.d/override.conf
|
||||
fi
|
||||
@@ -1,5 +1,6 @@
|
||||
#!/bin/bash
|
||||
|
||||
<<<<<<< HEAD
|
||||
if [ ! -d ./landing ]
|
||||
then
|
||||
mkdir landing
|
||||
@@ -49,6 +50,8 @@ then
|
||||
"
|
||||
fi
|
||||
|
||||
=======
|
||||
>>>>>>> version-1
|
||||
cat > ./landing/index.html <<EOL
|
||||
<html lang="en">
|
||||
|
||||
@@ -98,7 +101,18 @@ cat > ./landing/index.html <<EOL
|
||||
</tr>
|
||||
</thead>
|
||||
<tbody>
|
||||
<<<<<<< HEAD
|
||||
${LOCAL_SERVICES}
|
||||
=======
|
||||
<tr>
|
||||
<td>Bridgehead</td>
|
||||
<td>Reverse Proxy <a href="https://e260-serv-05/dashboard/">Traefik</a></td>
|
||||
</tr>
|
||||
<tr>
|
||||
<td>DKTK</td>
|
||||
<td><a href="https://e260-serv-05/dktk-localdatamanagement/fhir/">Blaze</a></td>
|
||||
</tr>
|
||||
>>>>>>> version-1
|
||||
</tbody>
|
||||
</table>
|
||||
</div>
|
||||
|
||||
@@ -15,40 +15,42 @@ for prerequisite in $prerequisites; do
|
||||
# TODO: Check for specific version
|
||||
done
|
||||
|
||||
echo "Checking site.conf"
|
||||
echo "Checking /etc/bridgehead-config/"
|
||||
|
||||
## Download submodule
|
||||
if [ ! -d "/etc/bridgehead-config/" ]; then
|
||||
echo "Please set up the site-config folder. Instruction are in the readme."
|
||||
exit 1
|
||||
else
|
||||
echo "Done"
|
||||
fi
|
||||
|
||||
echo "Checking /etc/bridgehead-config/site.conf"
|
||||
|
||||
#check if site.conf is created
|
||||
if [ ! -f site.conf ]; then
|
||||
if [ ! -f /etc/bridgehead-config/site.conf ]; then
|
||||
echo "Please create your specific site.conf file from the site.dev.conf"
|
||||
exit
|
||||
exit 1
|
||||
else
|
||||
echo "Done"
|
||||
fi
|
||||
|
||||
#Load site specific variables
|
||||
source site.conf
|
||||
source /etc/bridgehead-config/site.conf
|
||||
|
||||
if [ -z "$site_name" ]; then
|
||||
echo "Please set site_name"
|
||||
exit 1
|
||||
fi
|
||||
|
||||
echo "Checking site-config module"
|
||||
|
||||
## Download submodule
|
||||
if [ ! -d "site-config" ]; then
|
||||
echo "Please set up the site-config folder. Instruction are in the readme."
|
||||
exit
|
||||
else
|
||||
echo "Site configuration is already loaded"
|
||||
fi
|
||||
|
||||
#Check if a project is selected
|
||||
if [ -z "$project" ]; then
|
||||
echo "No project selected! Please add a Project in your local site.conf."
|
||||
exit
|
||||
fi
|
||||
|
||||
#check if project env is present
|
||||
if [ -d "site-config/${project}.env" ]; then
|
||||
echo "Please copy the tempalte from ${project} and put it in the site-config folder"
|
||||
if [ -d "/etc/bridgehead-config/${project}.env" ]; then
|
||||
echo "Please copy the tempalte from ${project} and put it in the /etc/bridgehead-config/ folder"
|
||||
exit 1
|
||||
else
|
||||
echo "Done"
|
||||
fi
|
||||
|
||||
echo "All prerequisites are met!"
|
||||
|
||||
85
nngm/docker-compose.yml
Normal file
85
nngm/docker-compose.yml
Normal file
@@ -0,0 +1,85 @@
|
||||
version: "3.7"
|
||||
|
||||
services:
|
||||
traefik:
|
||||
container_name: bridgehead-traefik
|
||||
image: traefik:2.4
|
||||
command:
|
||||
- --api.insecure=true
|
||||
- --entrypoints.web.address=:80
|
||||
- --entrypoints.websecure.address=:443
|
||||
- --providers.docker=true
|
||||
- --entrypoints.web.http.redirections.entrypoint.to=websecure
|
||||
- --entrypoints.web.http.redirections.entrypoint.scheme=https
|
||||
ports:
|
||||
- 80:80
|
||||
- 443:443
|
||||
- 8080:8080
|
||||
volumes:
|
||||
- ../certs:/tools/certs
|
||||
- /var/run/docker.sock:/var/run/docker.sock:ro
|
||||
extra_hosts:
|
||||
- "host.docker.internal:host-gateway"
|
||||
|
||||
### Does need to know the outside proxy to connect central components
|
||||
forward_proxy:
|
||||
container_name: bridgehead-squid
|
||||
image: ubuntu/squid
|
||||
environment:
|
||||
http_proxy: ${http_proxy}
|
||||
https_proxy: ${https_proxy}
|
||||
volumes:
|
||||
- "bridgehead-proxy:/var/log/squid"
|
||||
|
||||
## Needs internal proxy config
|
||||
landing:
|
||||
container_name: bridgehead-landingpage
|
||||
image: nginx:stable
|
||||
volumes:
|
||||
- ../landing/:/usr/share/nginx/html
|
||||
labels:
|
||||
- "traefik.enable=true"
|
||||
- "traefik.http.routers.landing.rule=PathPrefix(`/`)"
|
||||
- "traefik.http.services.landing.loadbalancer.server.port=80"
|
||||
- "traefik.http.routers.landing.tls=true"
|
||||
|
||||
|
||||
nngm-connector:
|
||||
container_name: bridgehead-nngm-connector
|
||||
image: "samply/share-client:nngm-feature-environmentPreconfiguration"
|
||||
environment:
|
||||
POSTGRES_PASSWORD: ${POSTGRES_PASSWORD}
|
||||
NNGM_MAGICPL_APIKEY: ${NNGM_MAGICPL_APIKEY}
|
||||
NNGM_MAINZELLISTE_APIKEY: ${NNGM_MAINZELLISTE_APIKEY}
|
||||
NNGM_CTS_APIKEY: ${NNGM_CTS_APIKEY}
|
||||
NNGM_CRYPTKEY: ${NNGM_CRYPTKEY}
|
||||
volumes:
|
||||
- "nngm-connector-logs:/usr/local/tomcat/logs"
|
||||
labels:
|
||||
- "traefik.enable=true"
|
||||
- "traefik.http.routers.nngm_connector.rule=PathPrefix(`/nngm-connector`)"
|
||||
- "traefik.http.services.nngm_connector.loadbalancer.server.port=8080"
|
||||
- "traefik.http.routers.nngm_connector.tls=true"
|
||||
|
||||
depends_on:
|
||||
- "nngm-connector-db"
|
||||
- "forward_proxy"
|
||||
ports:
|
||||
- 5005:5005
|
||||
restart: "always"
|
||||
|
||||
nngm-connector-db:
|
||||
container_name: bridgehead-nngm-connector-db
|
||||
image: "postgres:10.17"
|
||||
environment:
|
||||
POSTGRES_DB: "share_v2"
|
||||
POSTGRES_USER: "samplyweb"
|
||||
POSTGRES_PASSWORD: ${POSTGRES_PASSWORD}
|
||||
volumes:
|
||||
- "nngm-connector-db-data:/var/lib/postgresql/data"
|
||||
restart: "always"
|
||||
|
||||
volumes:
|
||||
nngm-connector-db-data:
|
||||
nngm-connector-logs:
|
||||
bridgehead-proxy:
|
||||
@@ -1,18 +1,29 @@
|
||||
#!/bin/bash
|
||||
### Note: Currently not complete, needs some features before useable for production
|
||||
|
||||
if [ $# -eq 0 ]; then
|
||||
echo "Please provide a Project as argument"
|
||||
exit 1
|
||||
fi
|
||||
|
||||
if [ $1 != "ccp" ] && [ $1 != "nngm" ] && [ $1 != "gbn" ]; then
|
||||
echo "Please provide a supported project like ccp, gbn or nngm"
|
||||
exit 1
|
||||
fi
|
||||
|
||||
export project=$1
|
||||
|
||||
source lib/functions.sh
|
||||
|
||||
if ! lib/prerequisites.sh; then
|
||||
log "Prerequisites failed, exiting"
|
||||
exit
|
||||
log "Validating Prerequisites failed, please fix the occurring error"
|
||||
exit 1
|
||||
fi
|
||||
|
||||
./lib/generate.sh
|
||||
|
||||
log "Starting bridgehead"
|
||||
|
||||
# TODO: Check $1 for proper values
|
||||
docker-compose -f $1/docker-compose.yml --env-file bridgehead-config/$1.env up -d
|
||||
docker-compose -f $1/docker-compose.yml --env-file /etc/bridgehead-config/$1.env up -d
|
||||
|
||||
log "The bridgehead should be in online in a few seconds"
|
||||
|
||||
@@ -2,7 +2,17 @@
|
||||
|
||||
source lib/functions.sh
|
||||
|
||||
if [ $# -eq 0 ]; then
|
||||
echo "Please provide a Project as argument"
|
||||
exit 1
|
||||
fi
|
||||
|
||||
if [ $1 != "ccp" ] && [$1 != "nngm"] && [ $1 != "gbn" ]; then
|
||||
echo "Please provide a supported project like ccp, gbn or nngm"
|
||||
exit 1
|
||||
fi
|
||||
|
||||
log "Stopping bridgehead"
|
||||
|
||||
# TODO: Check $1 for proper values
|
||||
docker-compose -f $1/docker-compose.yml --env-file bridgehead-config/$1.env down
|
||||
docker-compose -f $1/docker-compose.yml --env-file /etc/bridgehead-config/$1.env down
|
||||
|
||||
Reference in New Issue
Block a user